The Case for Genomics
DNA is the fundamental biological unit of data storage. It comprises long sequences of base pairs (bps), each of which represent one of four values (A, T, C or G), similar in the way binary digit (bit) data storage is used in computing. Different combinations of these base pairs encode the blueprints for constructing every molecular machine that life needs to function.
A genome is the complete set of DNA sequences in an organism and contains all of the instructions required for that organism to function, including embryogenesis, growth, responding to the environment, and healing from disease. The Human Genome – the complete map of all 23 large DNA sequences (chromosomes) that encodes our species – comprises a total of around 3 billion base pairs contained within the nuclei of each of our cells.
While this sequence of base pairs is virtually identical in every human, differentiating us from, say, a chimpanzee or a mouse, there are nonetheless subtle differences in each of our individual genomes that make us unique. Whole-genome sequencing, pioneered by the Human Genome Project, enables us to read a person’s individual genome and, among other things, identify differences from the average human genome. Such differences (mutations) are often associated with disorders and disease but can also be associated with other factors like disease resistance or sensitivity to an environmental perturbation like sunlight or exercise.
There are a multitude of ‘single-gene’ disorders, including cystic fibrosis and sickle cell anaemia, which can arise from mutation of just one point in a single gene. More commonly, however, disease results from a complex combination of genetic and environmental factors, as is the case for cancers, dementia and cardiovascular diseases – the leading causes of death in advanced economies. The ability to read the genomic profile of an individual, in conjunction with understanding their environment, is critical in helping us to predict the likelihood of these diseases occurring, as well as to identify individuals that may be predisposed to certain risk factors. This is critical to informing lifestyle choices, the design of built environments, and medical decisions to prevent, treat or cure certain conditions.
The study of genomics also has enormous benefits at a population level. By aggregating genomic profiles from across the population we can develop invaluable datasets to which advanced analytics and AI can be applied to develop a much more detailed understanding of the polygenetic causative factors, and thus potential treatments, of disease. This is particularly the case for rare genetic diseases, which require large datasets to find statistically significant correlations, which can identify causations.
Over the past two decades, dramatic advancements in DNA-sequencing technologies have massively reduced the time and cost required to sequence an entire human genome (now less than $1,000/genome), making it feasible to have all of our genomes sequenced as part of routine health care in the near future, as has recently been proposed by Genomics England, in the UK.
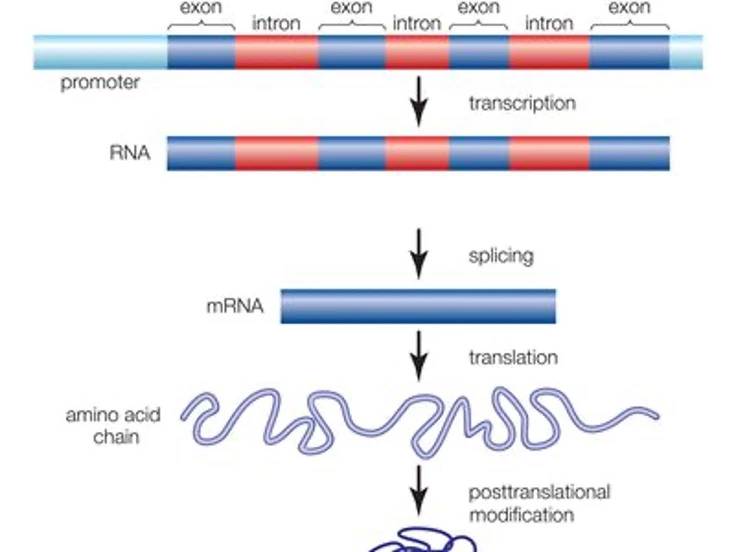
Figure 1 | Anatomy of genes and gene expression
https://www.britannica.com/science/gene
Genes and Genomes
The human genome contains around 25,000 genes, each of which typically encodes a single protein that performs a specific biological function in the cell, such as regulating cell metabolism, growth or shape.
In order to produce proteins, transcription factors, a family of proteins that directly interact with genomic DNA sequences, bind to DNA regions typically ‘upstream’ of a gene section known as a promoter. Gene expression is initiated by transcribing the gene sequence into complementary messenger RNA (mRNA), which then acts as a vehicle to transport this information from the nucleus into the main body of the cell. Here, the mRNA sequence is ‘translated’ via cellular machinery into a specific sequence of amino acids that then folds into a three-dimensional structure to form the intended protein. Prior to translation, mRNA can be ‘spliced’ to remove non-coding ‘introns’ (which have various regulatory, processing and evolutionary roles). Protein-coding regions, or ‘exons’, can also be spliced together in different combinations to produce variant protein forms (‘isoforms’) derived from the same gene.
Counterintuitively, however, DNA that encodes proteins (‘coding DNA’) only makes up around 1-2% of the human genome. The remainder is composed of non-coding DNA, including gene elements (e.g., introns, promoters etc.) and intergenic regions, which lie between genes. Up to 80% of non-coding DNA is now thought to have a biological function and the ENCODE project has mapped these elements across the human genome. The precise biological functions of many of these non-coding regions are still being determined, but it is thought that much of it contributes to gene regulation (where, when and to what extent a gene is expressed) and genome organisation.
Mutations in coding DNA, which can be inherited or accumulate throughout our lifetime, can cause changes in the amino acid sequence of the encoded protein, which may cause a complete or partial loss of protein function, resulting in a biological defect. More rarely, gain-of-function mutations can occur, which cause excessive protein activity, which too can lead to disease. Meanwhile, mutations in non-coding regions can potentially effect gene regulation (including affecting multiple genes simultaneously), which can also cause, or be a contributing factor, to disease. The interactions between these mutations across the landscape of the human genome are, however, complex, and still being mapped out, as specific differences in one person’s genome may have different effects than those in another person’s genome.
Understanding our own genomes then, particularly where and what mutations we have in coding or non-coding regions of DNA, is critical to understanding our personal molecular biology and our personal health. Sequencing the genomes of other species, meanwhile is useful in understanding the biology of commercial crops, pathogens causing infectious diseases and model organisms to understand fundamental concepts of biology.
Common Types of Sequencing
A range of different sequencing strategies have been developed that trade speed and cost against depth of comprehension:
Whole-genome sequencing - sequences the entire genome (including coding and non-coding regions) of an individual organism. The sequenced genome is then analysed against a reference genome for that species.
Whole-exome sequencing - sequences only the protein-coding regions (exons) of the genome. Exome sequencing and analysis is quicker than whole genome sequencing but will not elucidate mutations in non-coding DNA.
SNP genotyping – only sequences specific genetic loci where single base pair changes, also known as Single Nucleotide Polymorphisms (or SNPs – pronounced ‘snips’), are known to be associated with certain traits. Companies such as 23andMe use this approach to provide insights on genetic heritage and disease risk. This approach is also much cheaper and quicker than whole-genome sequencing but provides relatively, more limited information.
RNA sequencing – sequences complimentary DNA (cDNA) of each mRNA molecule in tissues or single cells, enabling quantification of gene expression which can provide insight into how a given cell type is operating.
Third-generation Sequencing
First-generation sequencing, also known as Sanger sequencing, was first developed in 1977 by British biochemist Fredrick Sanger. This technique, which utilises random termination and electrophoresis to read each “letter” of the genome, was used in the Human Genome Project, which took around 13 years to complete. It’s highly labour-intensive and expensive.
Sanger sequencing was superseded by Next Generation Sequencing (NGS; also known as high-throughput sequencing), which became commercially available from 2005. NGS uses a massively parallel synthesis reading technique to simultaneously read 1 million to 40 billion short strands of DNA (typically between 50-400 bps in length) in a single run. As a result, an entire human genome can be sequenced in a day, greatly reducing costs.
More recently a novel sequencing technology, first developed by Oxford Nanopore Technologies, has been dubbed ‘third generation’, It uses nanopores (nano-scale holes) embedded in high tech electronics to perform precise molecular analyses. It can sequence anything from short to ultra-long strands of DNA and RNA (500bps – 2Mbs), measure DNA methylation (which affects gene expression) and has the potential to be able to sequence amino acid sequences. Unlike previous sequencing techniques, genetic material does not need to be amplified before sequencing, making the process shorter and easier. Importantly, Oxford Nanopore has developed a number of sequencing devices that range from small portable devices to high-throughput desktop machines, which are coupled to a real-time data delivery platform.
There are several advantages to this technology:
It’s easier and quicker to prepare samples for sequencing
It can sequence DNA and RNA faster than NGS techniques – sequencing an entire Human genome in as little as just 5 hours
Ultra-long reads enable more accurate, and faster, genome assembly
Portable devices mean genomes can be sequenced in any setting
Real-time data read outs can help inform immediate medical decisions
It can be cheaper to use
Widespread adoption of this technology designed so ‘anyone anywhere can sequence anything’ has the potential to have major impacts on health care, epidemiology and AgTech. Making genomics sequencing and analysis faster, cheaper and more mobile has obvious benefits for health care, particularly in terms of real-time diagnosis for everything from cancer to infectious diseases. Meanwhile, portability and the immediacy of results has profound potential for real-time, high-resolution identification and monitoring of novel pathogens, both for human patients and in agriculture. Meaning we can rapidly detect the emergence of new diseases or disease strains in a highly precise manner, supporting efforts for widespread genomic surveillance of variants of concern for diseases like as Covid-19.
There are few technologies that promise to be as informative and transformative as genomics. From the improvements to personalised medicine and national and international public health policy to the economic gains generated by improved health and crop management, governments will be hard-pressed to find a better value for money proposition. Investment in national and regional genomics facilities should be a first order priority in advancing the future of health.
Acknowledgements
The authors would like to thank Dr. Alexander Eve for review.